Analysis of RNA kinetics
using

Alexey Uvarovskii, Christoph Dieterich
University Hospital Heidelberg
 Alexey Uvarovskii. pulseR for RNA kinetics
Alexey Uvarovskii. pulseR for RNA kinetics
Analysis of RNA kinetics
using

Alexey Uvarovskii, Christoph Dieterich
University Hospital Heidelberg


Tobias Jakobi (Dieterich Lab) - computing support
David Vilchez, Seda
Koyuncu (CECAD Cologne),
Janine Altmüller, Marek Franitza (CCG Cologne) -
experimental data
Number of motor vehicle deaths in the US (wiki)
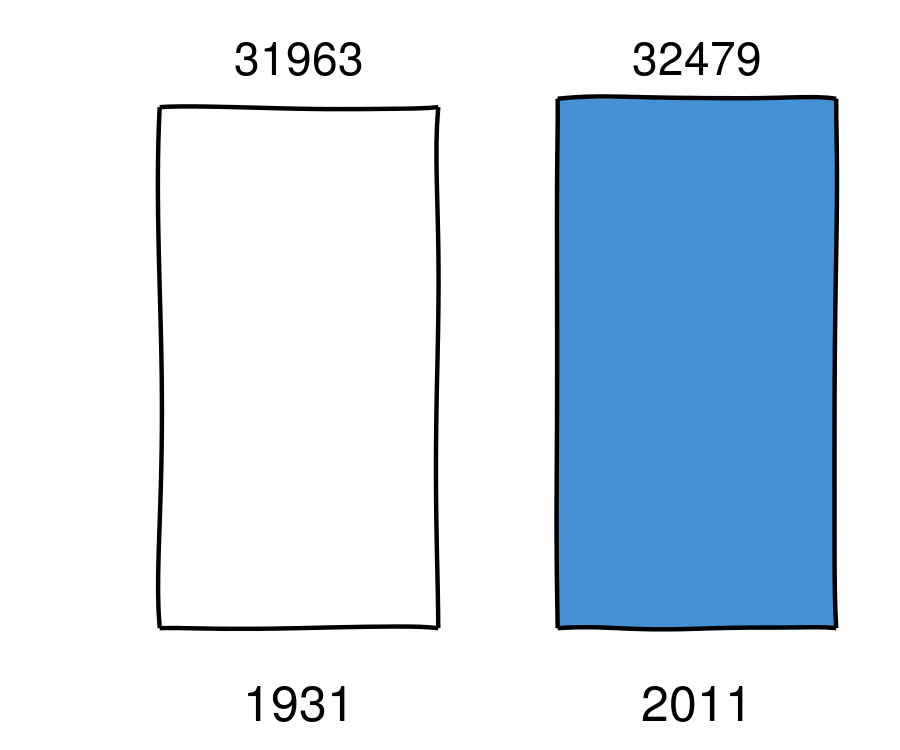
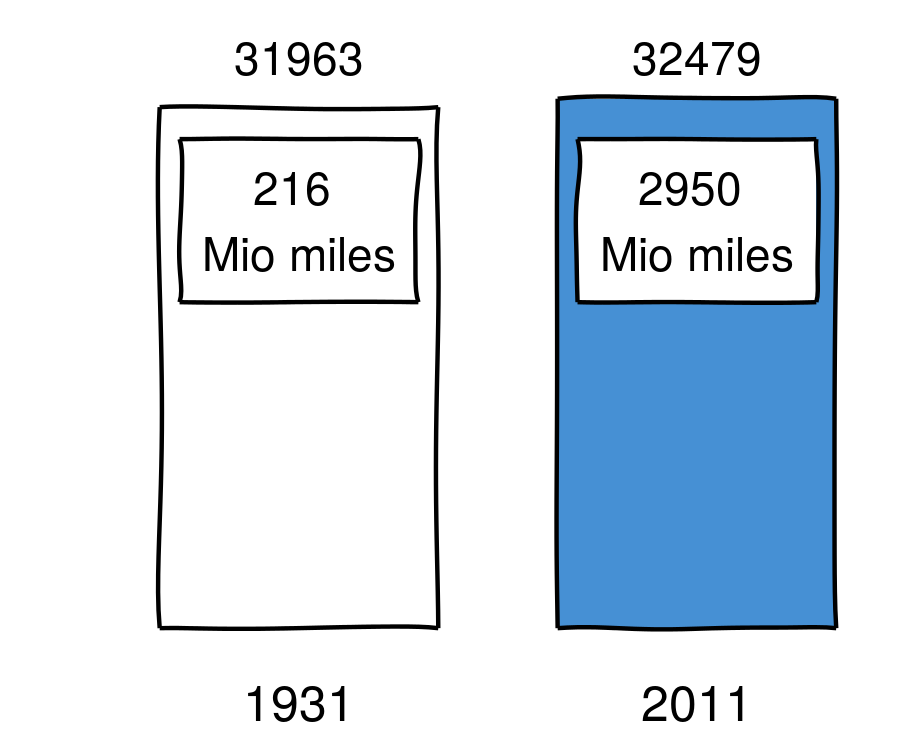
Number of motor vehicle deaths in the US (wiki)

$\sf \frac{d[\text{RNA}]}{dt} = +$ $\sf [\text{synthesis}] $ $\sf -$ $\sf [\text{degradation}]$ $\sf \cdot [\text{RNA}]$
$$ \sf [\text{steady state RNA}] = \frac{[\text{synthesis}]}{[\text{degradation }]}$$
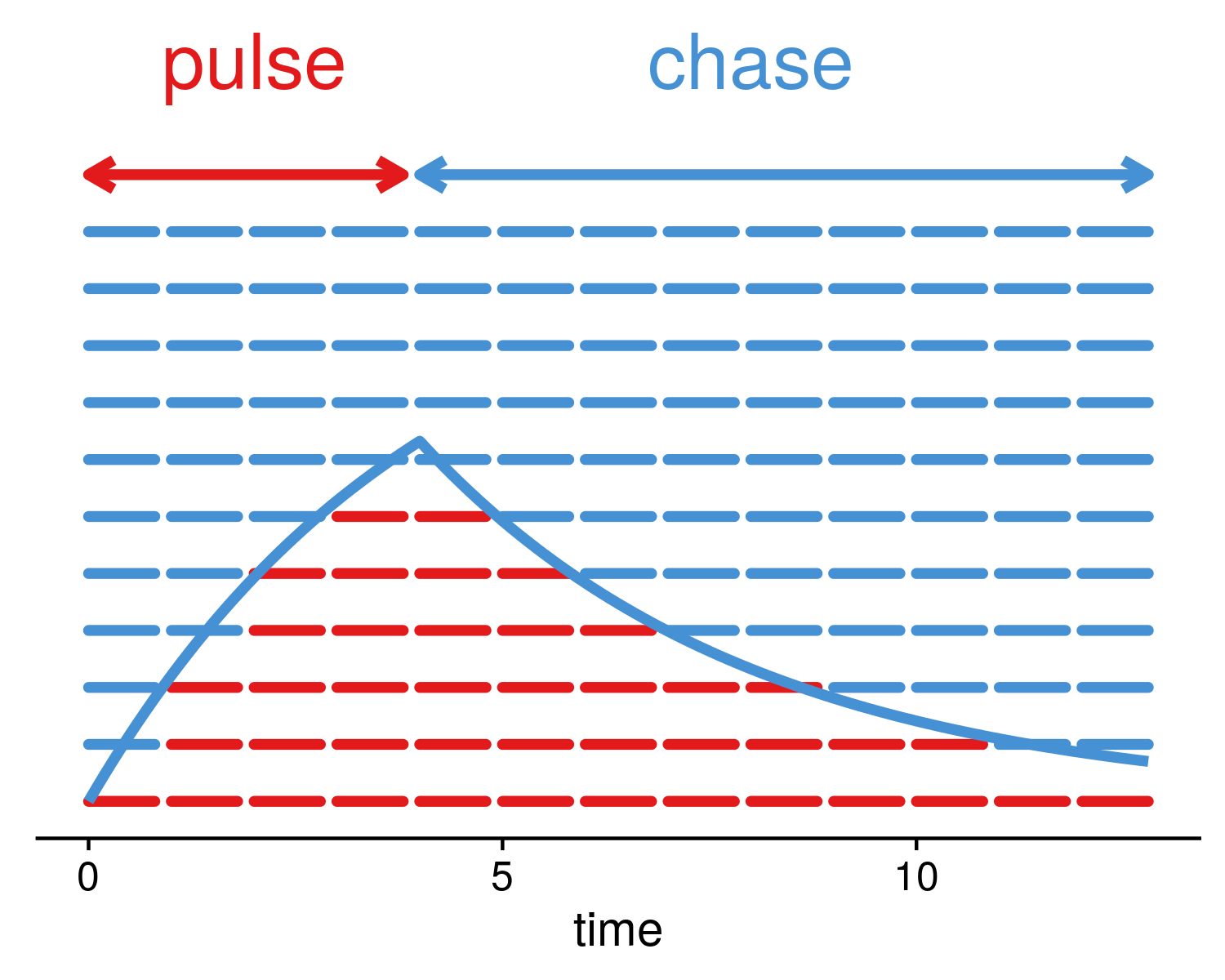
EU = 5-ethyniluridine
a way to measure RNA kinetics
Alexey Uvarovskii, Christoph Dieterich; pulseR: Versatile computational analysis of RNA turnover from metabolic labeling experiments. Bioinformatics 2017 btx368. doi: 10.1093/bioinformatics/btx368
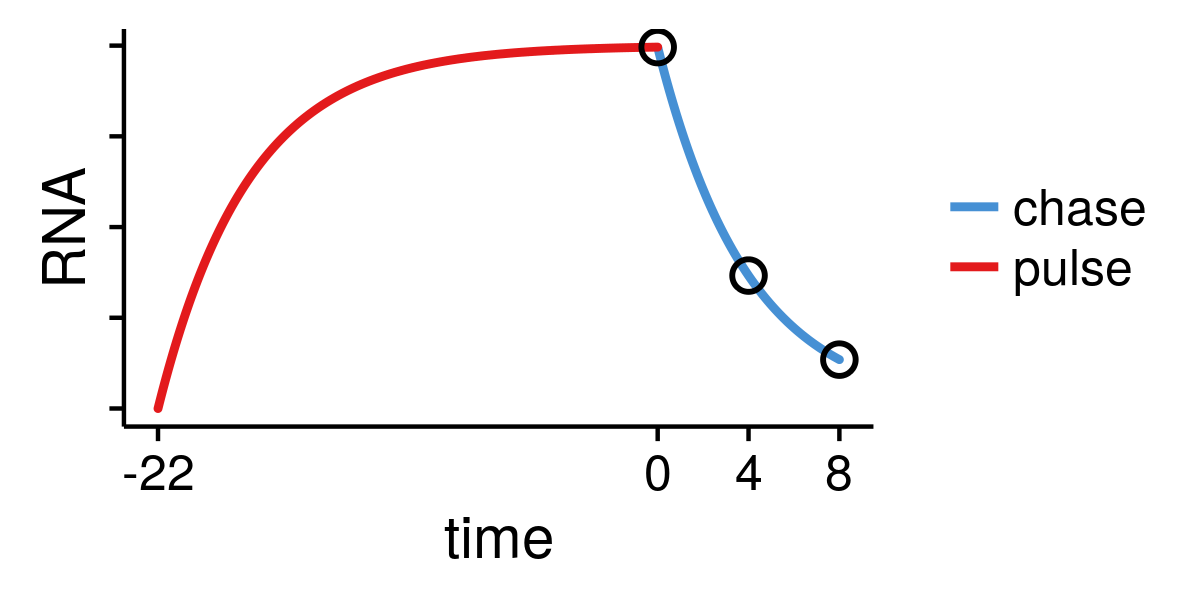
defined by the setup, e.g. pulse labelling is
$$ \begin{align} \sf [\text{total}] &= T\equiv\text{const} \\ \sf[\text{pull down}] &= T\cdot \left( 1 - e^{-dt}\right) \end{align}$$
$$\sf t = 1, 2, 4\, \text{hr} $$
Negative binomial distribution
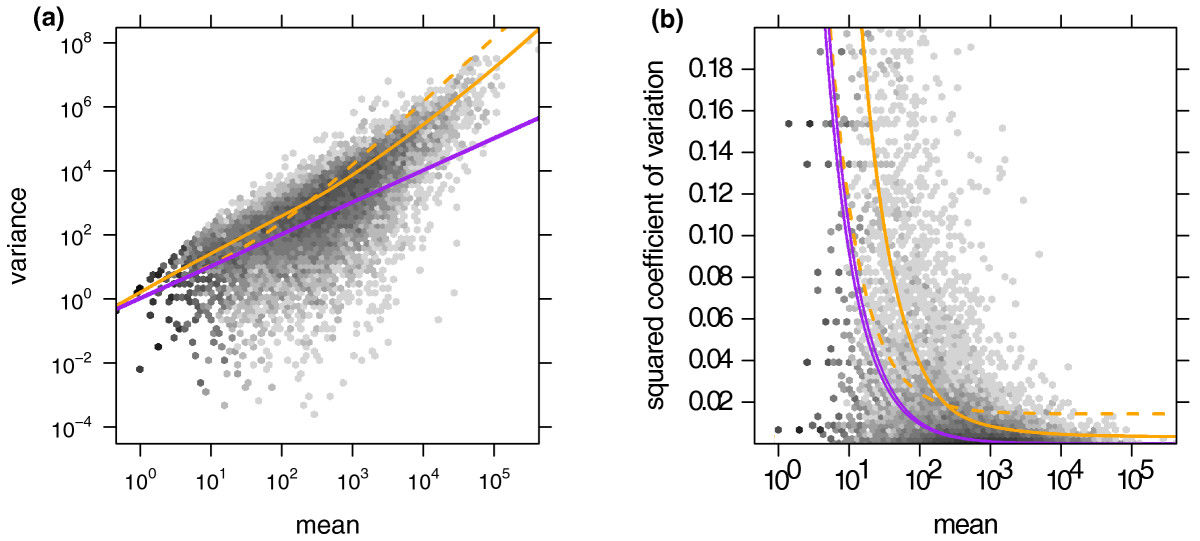
purple: no overdispersion, yellow: with overdispersion
Anders, Simon, and Wolfgang Huber. "Differential expression analysis for sequence count data." Genome biology 11.10 (2010): R106.
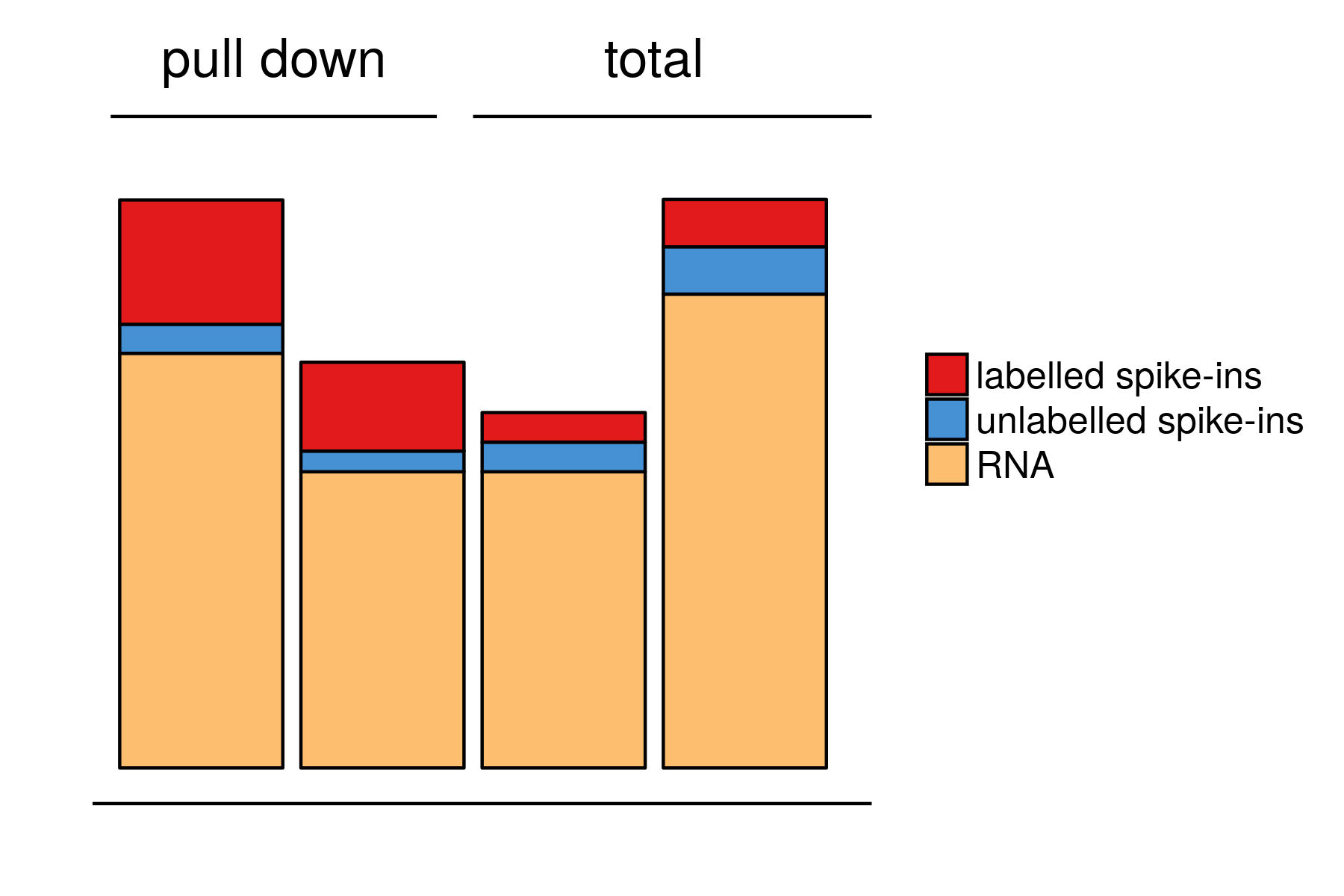
using spike-ins (DESeq)
by MLE fitting
absolute synthesis rate
no spike-ins needed
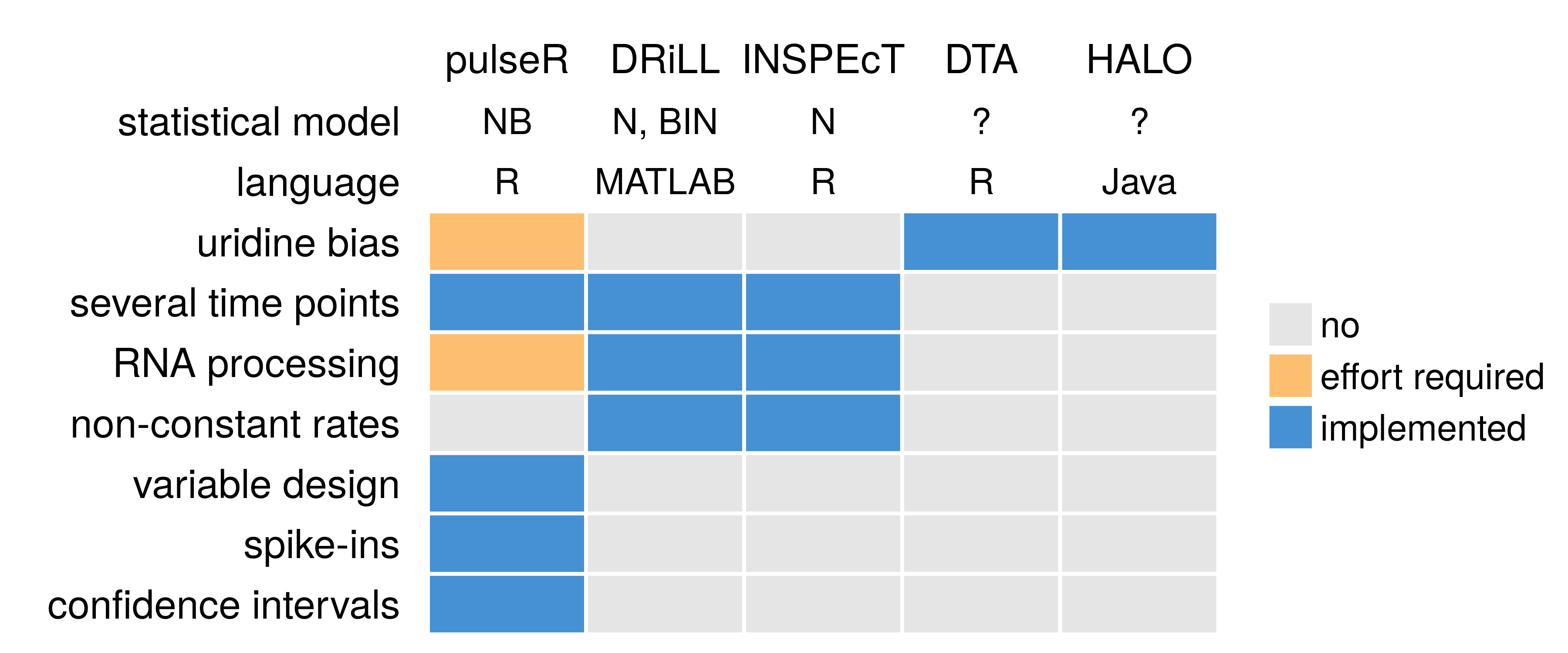
N: normal, NB: negative binomial, BIN: binomial.

design
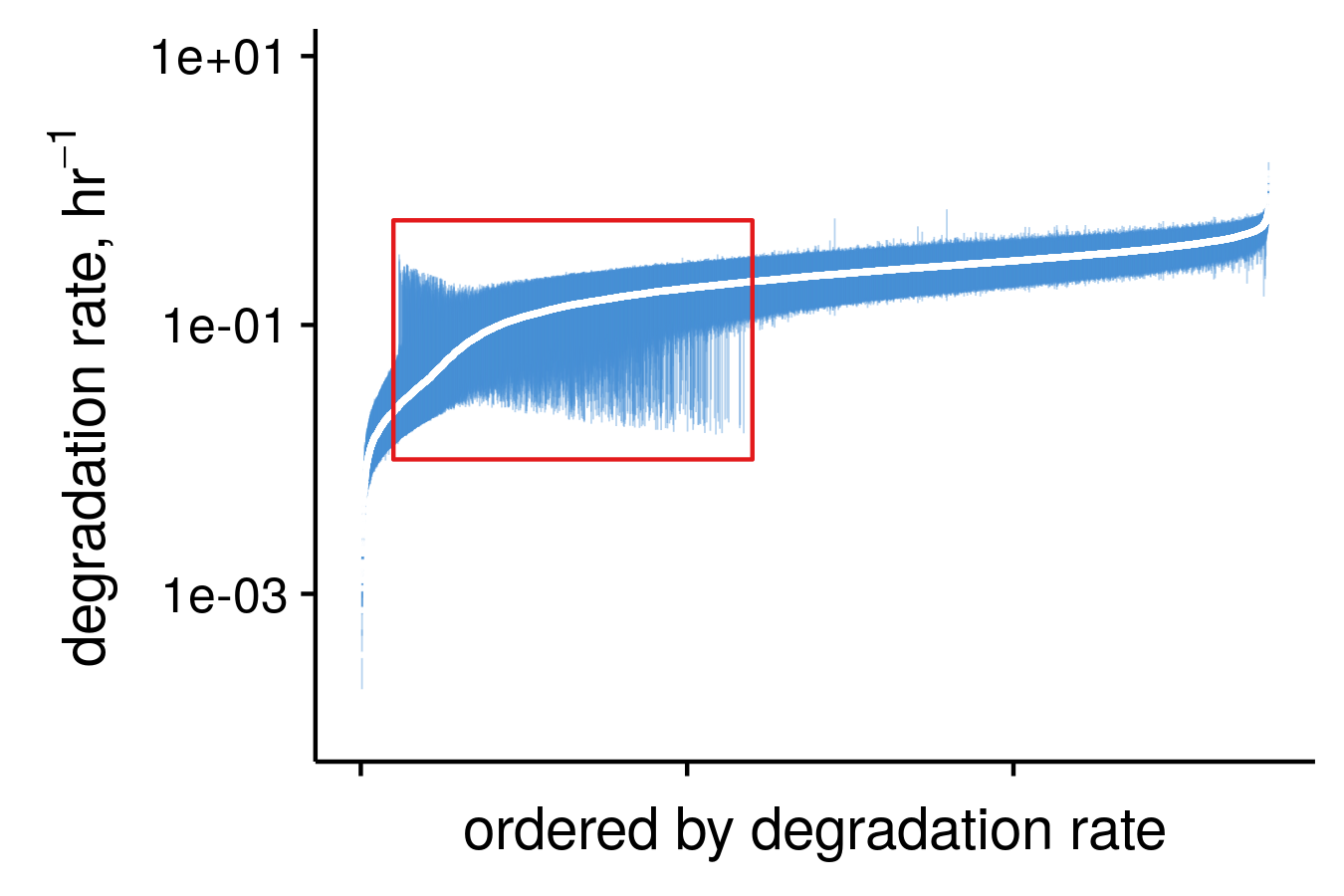
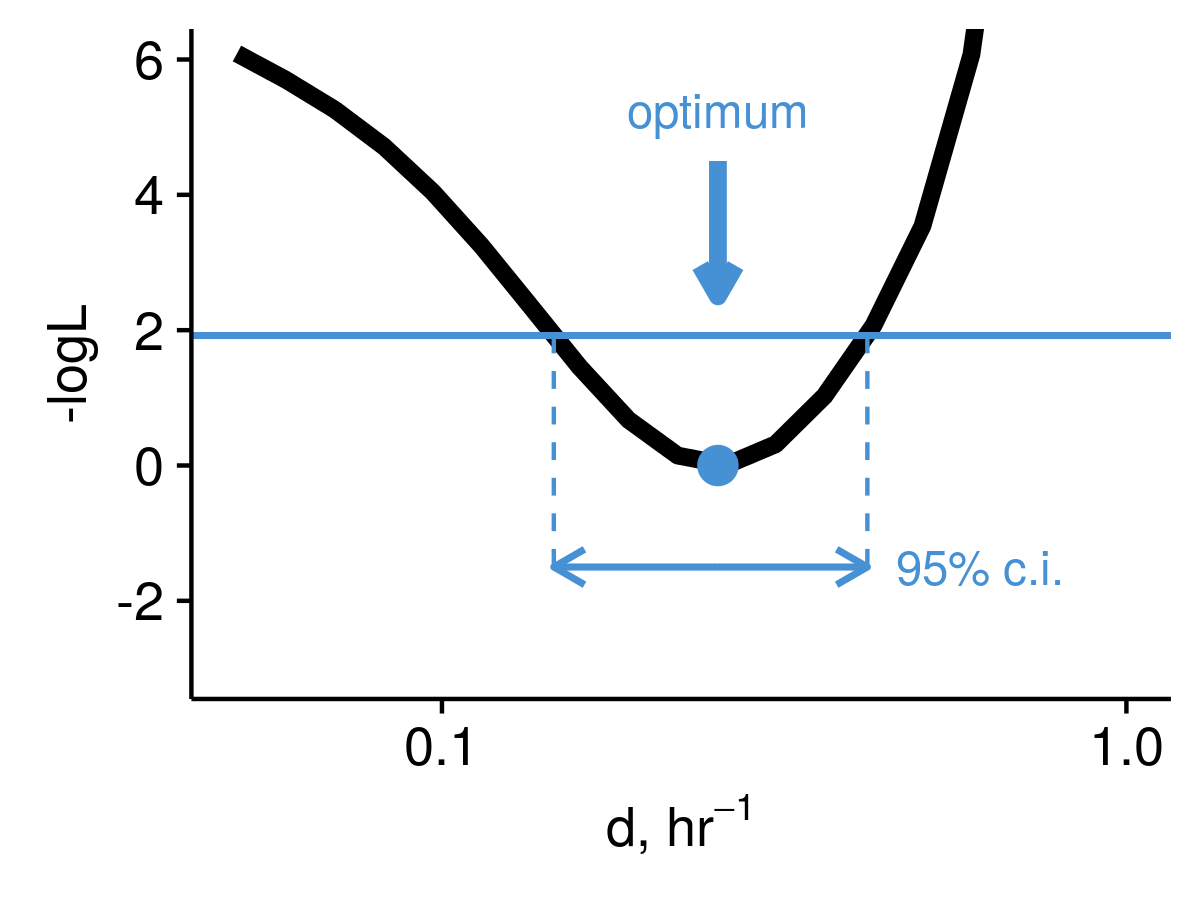
pulseR can estimate confidence intervals for you
diagnostics and comparisons

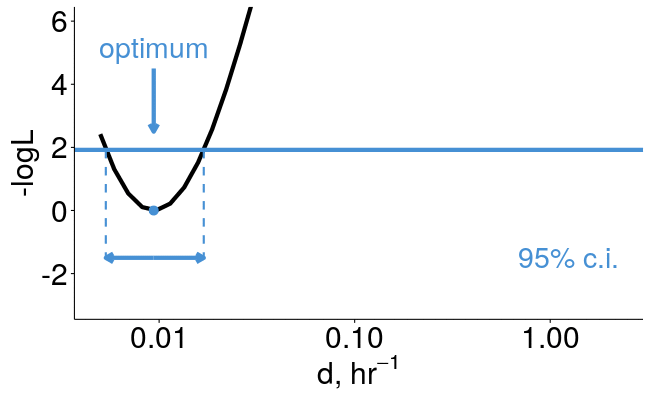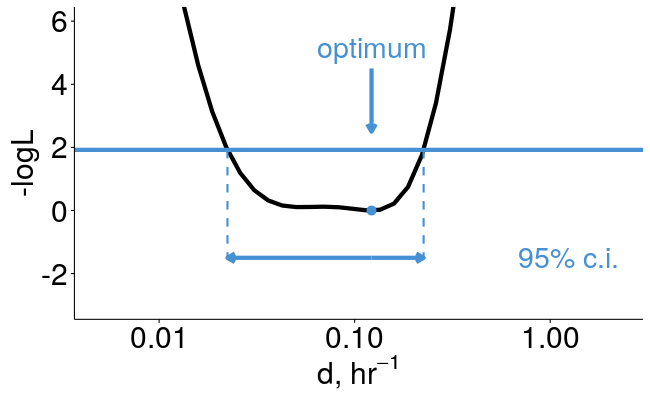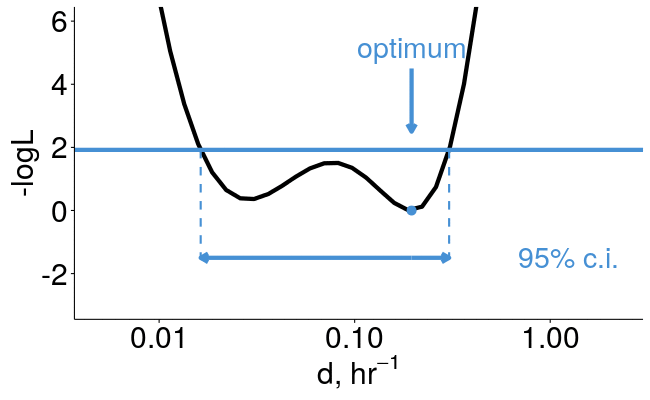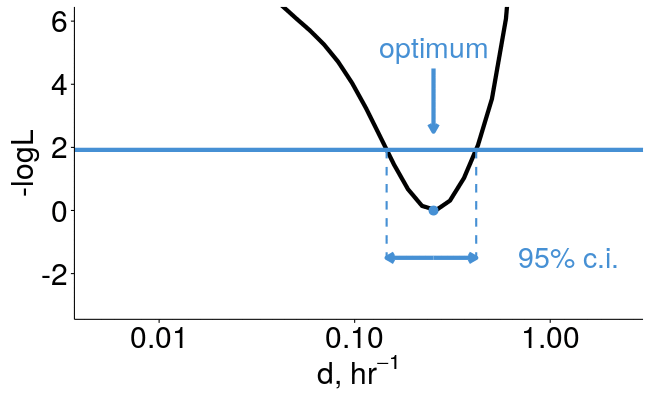
library(pulseR)
# put math here
formulas <- MeanFormulas(
total = mu,
labelled = mu * (1 - exp(-d*22)) * exp(-d*time),
unlabelled = mu * (1 - exp(-d*time) * (1 - exp(-d * 22)))
)
# define the fractions
formulaIndexes <- list(
total_fraction = 'total',
pull_down = c('labelled', 'unlabelled'))
pd <- PulseData(counts, conditions, formulas, formulaIndexes,
groups = ~ fraction + time)
result <- fitModel(pd, initValues, opts)
Poster A-271
An open post-doc position


a.uvarovskii@uni-heidelberg.de